Welcome!
We explore the ways in which natural selection and evolutionary change have shaped the human species (and other mammals too!). By combining the versatility of induced pluripotent stem cells with the power of functional genomics we address questions about the mechanisms of evolutionary adaptation in humans that are intractable by any other means.
We are part of the Melbourne Integrative Genomics unit and the School of BioSciences at the University of Melbourne, and have very strong connections to the Centre for Stem Cell Systems. We are also an affiliate member of Stem Cells Australia, a multi-institution, multi-disciplinary initiative to bring together Australia’s leading stem cell research groups.
We are always looking for enthusiastic people to join the lab. If you are interested in pursuing an Masters or PhD degree with us, contact Irene and we’ll go from there.
The lab is focused on human evolution, especially in the ways in which gene regulatory processes have contributed to it. Because gene regulation is not easily predicted (at least for now!) from sequence-level data, we use induced pluripotent stem cells (iPSCs) as models to understand the means by which multiple cellular mechanisms interact to regulate gene expression, and, under the action of natural selection, ultimately give rise to inter-species or population-level differences. We are one of few labs worldwide with an established record in generating iPSC lines from non-model organisms, and we combine this expertise with our experience working with the latest genomic technologies.
At the University of Melbourne we enjoy very close links with the School of BioSciences and of Mathematics and Statistics, as well as the Centre for Stem Cell Systems, and we are affiliate members of Stem Cells Australia, a multi-institution, multi-disciplinary initiative that brings together Australia’s leading stem cell research groups across different expertises and interests. Further afield, we also have links to researchers at institutions like MCRI, A*STAR, Massey University, Nanyang Technological University, Monash Malaysia, the Wellcome Trust Sanger Institute, Penn State University, the Centro de Regulación Genómica in Barcelona and more.
Our lab is committed to carrying out high quality reproducible research, be that computational or molecular biology based. To that end we make source code and all data and protocols available on publication
Below you can find brief descriptions of some of our research interests.
The role of gene regulatory changes in human evolution
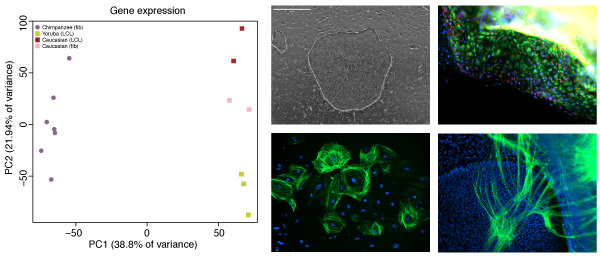
The majority of DNA differences between humans and chimpanzee lie outside protein coding regions of the genome, strongly arguing for a large role of gene regulation as a driver of evolutionary change, especially during embryonic development. However, our understanding of the mechanisms of gene regulation is far from complete, and without access to the right tissue type, it is very difficult to predict the consequences of any mutation. Working with iPSCs gives us access to the right cell type, at the right developmental time point, from the right animal, and enables us to understand the role these differences play in giving rise to particular phenotypes.
Key publications:
Gallego Romero*, Pavlovic* et al, eLife, 2015.
Gallego Romero, Gilad and Ruvinski, Nat Rev Genetics, 2012.
Local adaptations in human populations
The iPSC-driven comparative evolutionary paradigm can also be pertinent to questions about local adaptation in human groups. Of particular interest to us are populations living in the Arctic Circle, who have had to adapt to life in extreme cold conditions, populations adapted to life at high altitude in both Asia and South America, as well as populations living in Island South East Asia, who have been traditionally under-studied. Although there is well-documented evidence for local adaptation in all cases, the functional mechanisms remain elusive. As above, our goal is to combine readily available allelic and sequence information with iPSCs generated from these groups and control populations in order to investigate the actual consequences of these sequence differences in living cells. However, our focus here is not the identification of developmental differences between groups, but rather the identification of population-level functional differences in terminal cell types.
Key publications:
Gallego Romero et al, MBE, 2011.
Metspalu*, Gallego Romero*, Yunushbaev* et al, AJHG, 2011.
Better methods for complex RNA-seq
The use of RNA-seq is widespread. But many experiments – those that depend on field samples, or limited input, or atypical comparisons, suffer from biases that are not often discussed or explored at great depth. We are interested in quantifying and correcting for the effect of these confounders, and in developing analytical approaches that can attenuate these effects. Presently we are most keenly interested in developing robust methods to extensively and affordably perform inter-species RNA-seq comparisons.
Key publications:
Gallego Romero et al, BMC Biology, 2014.
Current members:

Isabela Alvim
Postdoctoral researcher
isabelaalvim dot alvim at unimelb dot edu do au
Isobel Beasley
MSc student, Genetics, co-supervised with Christina Azodi.

Maddy Comerford
PhD student, Genetics
mlcomerford at student dot unimelb dot edu dot au
Kei Fukuda
Postdoctoral researcher

Irene Gallego Romero, PhD
Principal investigator
irene [dot] gallego [at] unimelb [dot] edu [dot] au 
![]()
![]()
I’m a human evolutionary biologist with training in genomics, biological anthropology and pluripotent stem cell biology. My research interests include understanding evolutionary adaptions at both short and long time spans – those that have occurred in specific human populations, but also those that separate us from our close evolutionary relatives. I’m also fascinated by issues surrounding the practice of science today – funding allocation, career progression and levels of inclusion and diversity of the scientific workforce. You can download my current CV here (current as of Nov 2021).
I was a speaker at TEDxNTU in 2016. You can watch the recording here.
Hanadi Hoblos
Lab manager

Neke Ibeh
PhD student
oibeh [at] student [dot] unimelb [dot] edu [dot] au
Neke is applying her considerable bioinformatics skills to various genetic sets from Indonesia.

Navya Shukla
PhD student, Genetics
Joining the lab:
If you are looking for an honours, masters or PhD position, email me in the first instance, and we can chat to see if there’s any suitable positions available. You should also read through our lab guidelines document to get a feel for the lab and whether it is the right environment for you.
Past members:

Laura Cook
PhD student, co-supervised with Andrew Pask. (2018-2022)
Currently a postdoctoral researcher in the lab of Axel Visel at Berkeley National Labs.

Ashlee Hutchinson
PhD student, co-supervised with Andrew Pask. (2018-2022)
Currently a postdoctoral researcher in the lab of Robin Hobbs at Monash.

Davide Vespasiani
PhD student (2018-2022)
Currently a postdoctoral researcher in the lab of Carolyn De Graaf at WEHI

Katalina Bobowik
PhD student (2017-2021)
Currently a project scientist with Daniel MacArthur at the Garvan Institute.
Nicolas Brucato
Visiting postdoctoral research fellow (2019, 2020)
Debbie Lee Boon Hui
Undergraduate researcher (2020)
Jenna Ortoleva
Undergraduate researcher (2020)
River Kano
Masters in Biological Sciences (2018-2019)
Akihito Tomida
Research assistant (2019)
María Miguel Matorra
Undergraduate researcher (2019)
Anna Orteu
Research student (2017-2018)
And… back from our days in Singapore!
Pallavi Srivastava
Project officer/lab manager.
Ng Yan Ting
Project officer.
Leo Han Yun Sylvia
Final Year Project Undergraduate researcher.
Tan Wen Ting
Final Year Project Undergraduate researcher.
Phua Zi Jin Cheryl
Undergraduate URECA researcher.
Nguyen Thuan Tihn Sophia
CN Yang undergraduate scholar
Preprints
K. Bobowik, C. C. Darusallam, P. Kusuma, H. Sudoyo, C. A. Febinia, S. G. Malik, C. Wells and I. Gallego Romero, The whole blood microbiome of Indonesians reveals that environmental differences shape immune gene expression signatures, bioRxiv, doi: 10.1101/2022.04.24.489025. Full text available
I. Gallego Romero and A. Lea, Leveraging massively parallel reporter assays for evolutionary questions. arXiv: arXiv:2204.05857. Full text available
K. Bobowik, D. Syafruddin, C. C. Darusallam, H. Sudoyo, C. Wells and I. Gallego Romero, Transcriptomic profiles of Plasmodium falciparum and Plasmodium vivax-infected individuals in Indonesia. bioRxiv, doi: 10.1101/2021.01.07.425684. Full text available.
I. Gallego Romero, S. Gopalakrishnan and Y. Gilad, Widespread conservation of chromatin accessibility patterns and transcription factor binding activity in human and chimpanzee induced pluripotent stem cells. bioRxiv, doi: 10.1101/466631. Full text available.
2022
D. Vespasiani, G.S. Jacobs, L.E. Cook, N. Brucato, F.X. Ricaut, M. Leavesley, C. Kinipi, M.P. Cox and I. Gallego Romero. Denisovan introgression has shaped the immune system of present-day Papuans. PLOS Genetics, in press. Also available in bioRxiv, doi: 10.1101/2020.07.09.196444. Full text available.
H. M. Natri, G. Hudjashov, G. S. Jacobs, P. Kusuma, L. Saag, C C. Darusallam, M. Metspalu, H. Sudoyo, M. P. Cox*, I. Gallego Romero* and N. E. Banovich*, Genetic architecture of gene regulation in Indonesian populations identifies QTLs associated with local ancestry and archaic introgression. American Journal of Human Genetics, in press. Also available at bioRxiv, doi: 10.1101/2020.09.25.313726. * denotes equal contribution. Full text available.
2021
N. Shukla, B. Shaban and I. Gallego Romero, Genetic diversity in chimpanzee transcriptomics does not represent wild populations. Genome Biology and Evolution, in press. Also available at bioRxiv, doi:10.1101/2021.06.27.450107.
N. Brucato, M. André, R. Tsang, L. Saag, J. Kariwiga, K. Sesuki, T. Beni, W. Pomat, J. Muke, V. Meyer, A. Boland, J.F. Deleuze, H. Sudoyo, M. Mondal, L. Pagani, I. Gallego Romero, M. Metspalu, M.P. Cox, M. Leavesley and F.X. Ricaut, Papua New Guinean Genomes Reveal the Complex Settlement of North Sahul . Molecular Biology and Evolution doi:10.1093/molbev/msab238. Full text available.
M. Dannemann and I. Gallego Romero. Harnessing pluripotent stem cells as models to decipher human evolution. The FEBS Journal, doi:10.1111/febs.15885. Full text available.
2020
H. Natri*, K. S. Bobowik*, P. Kusuma, C. C. Darusallam, G. S. Jacobs, G. Hudjashov, J.S. Lansing, H. Sudoyo, N.E. Banovich**, M.P. Cox**, I. Gallego Romero**. Genome-wide DNA methylation and gene expression patterns reflect genetic ancestry and environmental differences across the Indonesian archipelago. PLOS Genetics, 16(5): e1008749. doi:10.1371/journal.pgen.1008749. * and ** denote equal contributions. Full text available.
K.X. Lau, E.A. Mason, J. Kie, D.P. De Souza, J. Kloehn, D. Tull, M.J. McConville, A. Keniry, T. Beck, M.E. Blewitt, M.E. Ritchie, S.H. Naik, D. Zalcenstein, O. Korn, S. Su, I Gallego Romero, C. Spruce, C.L. Baker, T.C. McGarr, C.A. Wells, and M.F. Pera. Unique Properties of a Subset of Human Pluripotent Stem Cells With High Capacity for Self-Renewal. Nature Communications, doi: 10.1038/s41467-020-16214-8.
2019
B. Yelmen, M. Mondal, D. Marnetto, A.K. Pathak, F. Montinaro, I. Gallego Romero, T. Kivisild, M. Metspalu and L. Pagani. Ancestry-specific analyses reveal differential demographic histories and opposite selective pressures in modern South Asian populations. Molecular Biology and Evolution, doi: 10.1093/molbev/msz037. Full text available.
2017
N. Banovich, Y. Li, A. Raj, M.C. Ward, P. Greenside, D. Calderon, P.Y. Tung, J.E. Burnett, M. Myrthil, S.M. Thomas, C.K. Burrows, I. Gallego Romero, B.J. Pavlovic, A. Kundaje, J.K. Pritchard and Y. Gilad. Impact of regulatory variation across human iPSCs and differentiated cells. Genome Research, 2017. doi:10.1101/gr.224436.117. Full text available.
2016
S. Mallick, H. Li*, M. Lipson*, I. Mathieson*, M. Gymrek, F. Racimo, J.P. Spence, M. Zhao, N. Chennagiri, S. Nordenfelt, A. Tandon, P. Skoglund, I. Lazaridis, S. Sankararaman, Q. Fu, N. Rohland, G. Renaud, Y. Erlich, T. Willems, C. Gallo, G. Poletti, F. Balloux, G. van Driem, P. de Knifjj, I. Gallego Romero, A.R. Jha, D.M. Behar, C.M. Bravi, C. Capelli, T. Hervig, A. Moreno- Estrada, O.L. Posukh, E. Balanovska, O. Balanovsky, S. Karachanak-Yankova, H. Sahakyan, D. Toncheva, L. Yepiskoposyan, C. Tyler-Smith, Y. Xue, M.S. Abdullah, A. Ruiz-Linares, C.M. Beall, A. Di Rienzo, C. Jeong, E.B. Starikovskaya, E. Metspalu, J. Parik, R. Villems, B.M. Henn, U. Hodoglugil, R. Mahley, A. Sajantila, G. Stamatoyannopoulos, J.T. S. Wee, R. Khusainova, E. Khusnutdinova, S. Litvinov, G. Ayodo, D. Comas, M. Hammer, T. Kivisild, W. Klitz, C. Winkler, D. Labuda, M. Bamshad, L.B. Jorde, S.A. Tishko , W.S. Watkins, M. Metspalu, S. Dryomov, R. Sukernik, L. Singh, K. Thangaraj, Y.S. Song, S. Pääbo, J. Kelso, N. Patterson, and D. Reich. The landscape of human genome diversity. Nature, doi:10.1038/nature18964. * denotes equal contribution. Full text available.
C.L. Kagan*, N.E. Banovich*, B.J. Pavlovic, K. Patterson, I. Gallego Romero, J.K. Pritchard and Y. Gilad. Genetic Variation, Not Cell Type of Origin, Underlies Regulatory Differences in iPSCs. PLoS Genetics, doi: 10.1371/journal.pgen.1005793. Also available at bioRxiv, doi: 10.1101/013888. * denotes equal contribution. Full text available.
2015
I. Gallego Romero*, B.J. Pavlovic*, I. Hernando-Herraez, X. Zhou, M.C. Ward, N.E. Banovich, C.L. Kagan, J.E. Burnett, C.H. Huang, A. Mitrano, C.I. Chavarria, I.F. Ben-Nun, Y. Li, K. Sabatini, T.R. Leonardo, M. Parast, T. Marques-Bonet, L..C. Laurent, J.F. Loring, and Y. Gilad. Generation of a Panel of Induced Pluripotent Stem Cells From Chimpanzees: a Resource for Comparative Functional Genomics. eLife, doi: 10.7554/eLife.07103. Also available at bioRxiv, doi: 10.1101/008862. * denotes equal contribution. Full text available.
P.H. Sudmant, S. Mallick, B.J. Nelson, F. Hormozdiari, N. Krumm, J. Huddleston, B.P. Coe, C. Baker1, S. Nordenfelt, M. Bamshad, L.B. Jorde, O.L. Posukh, H. Sahakyan, W.S. Watkins, L. Yepiskoposyan, M.S. Abdullah, C.M. Bravi, C. Capelli, T. Hervig, J.T.S. Wee, C. Tyler-Smith, G. van Driem, I. Gallego Romero, A.R. Jha, S. Karachanak-Yankova, D. Toncheva, D. Comas, B. Henn, T. Kivisild, A. Ruiz-Linares, A. Sajantila, E. Metspalu, J. Parik, R. Villems, E.B. Starikovskaya, G. Ayodo, C. Beall, A. Di Rienzo, M. Hammer, R. Khusainova, E. Khusnutdinova, W. Klitz, C. Winkler, D. Labuda, M. Metspalu, S.A. Tishkoff, S. Dryomov, R. Sukernik, N. Patterson, D. Reich, and E.E. Eichler. Global diversity, population stratification, and selection of human copy number variation. Science, doi: 10.1126/science.aab3761. Full text available.
I. Garitaonandia*, H. Amir*, F.S. Boscolo*, G.K. Wambua, H.L. Schultheisz, K. Sabatini, R. Morey, S. Waltz, Y-C. Wang, H. Tran, T.R. Leonardo, K. Nazor, I. Slavin, C. Lynch, Y. Li, R. Coleman, I. Gallego Romero, G. Altun, D. Reynolds, S. Dalton, M. Parast, J.F. Loring** and L.C. Laurent**. Increased Risk of Genetic and Epigenetic Instability in Human Embryonic Stem Cells Associated with Specific Culture Conditions. PLOSone, doi: 10.1371/journal.pone.0118307. * and ** denote equal contributions. Full text available.
2014
I. Gallego Romero, A. A. Pai, J. Tung and Y. Gilad. RNA-seq: Impact of RNA degradation on transcript quantification. BMC Biology, 12:42 doi:10.1186/1741-7007-12-42. Full text available.
A. Cardona, L. Pagani, T. Antão, D.J. Lawson, C.A. Eichstaedt, B. Yngvadottir, M. Than Than Shwe, J. Wee, I. Gallego Romero, S. Raj, M. Metspalu, R. Villems, E. Willerslev, C. Tyler-Smith, B.A. Malyarchuk, M.V, Derenko and T. Kivisild. Genome-wide analysis of cold adaption in indigenous Siberian populations. PLOSone, doi: 10.1371/journal.pone.0098076. Full text available.
I. Lazaridis, N. Patterson, A. Mittnik, G. Renaud, S. Mallick, P.H. Sudmant, J. Schraiber, S. Castellano, K. Kirsanow, C. Economou, R. Bollongino, K.I. Bos, S. Nordenfelt, C. de Filippo, K. Pruefer, A. Tandon, S. Sawyer, C. Posth, G. Ayodo, H.M.A. Babiker, E. Balanovska, O. Balanovsky, C. Beall, H. Ben-Ami, J. Bene, F. Berrada, F. Brighelli, G. Busby, F. Cali, C. Capelli, M. Churnosov, D.E.C. Cole, L. Damba, G. van Driem, S. Dryomov, S. Fedorova, M. Francken, I. Gallego Romero, X. Gebremedhin, M. Gubina, M. Hammer, B. Henn, U. Hodoglugil, A. Jha, R. Kittles, E. Khusnutdinova, T. Kivisild, V. Kučinskas, R. Kusainova, L. Kushniarevich, L. Laredj, S. Litvinov, T. Loukidis, R. Mahley, B. Melegh, E. Metspalu, T. Moen, J. Mountain, T. Nyambo, L. Ossipova, J. Parik, F. Platanov, O. Posukh, A. Di Rienzo, V. Romano, I. Rudan, R. Ruizbakiev, H. Sahakyan, A. Salas, M. Shriver, A. Tarekegn, D. Toncheva, S. Turdikulova, I. Uktveryte, O. Utevska, M. Voevoda, J. Wahl, P. Zalloua, L. Yeppiskoposyan, T. Zemunik, M. Thomas, S. Tishkoff, R. Villems, D. Comas, L. Singh, K. Thangaraj, R. Sukernik, M. Metspalu, M. Meyer, E.E. Eichler, J. Burger, M. Slatkin, S. Pääbo, J. Kelso, D. Reich, and J. Krause. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature, doi: 10.1038/nature13673. Also available at bioRxiv, doi: 10.1101/001552. Full text available.
2013
A. Bamberg Migliano, I. Gallego Romero, M. Metspalu, M. Leavesley, L. Pagani, T. Antão, D.W. Huang, B.T. Sherman, K. Siddle, C. Scholes, G. Hudjashov, E. Kaitokai, H. Mandui, A. Babalu, M. Belatti, A. Cagan, B. Hopkinshaw, C. Shaw, K. Tabbada, M. Nelis, E. Metspalu, R. Mägi, R.A. Lempicki, R. Villems, M. Mirazón Lahr and T. Kivisild. Evolution of the pygmy phenotype: evidence of positive selection from genome-wide scans in African, Asian and Melanesian pygmies. Human Biology, 85(1-3):251-84. Link.
C. Basu Mallick*, F.M. Iliescu*, M. Möls, S. Hill, R. Tamang, G. Chaubey, R. Goto, S.Y.W Ho, I. Gallego Romero, F. Crivellaro, G. Hudjashov, N. Rai, M. Metspalu, C.G.N. Mascie-Taylor, R. Pitchappan, L. Singh, M. Mirazón Lahr, K. Thangaraj, R. Villems and T. Kivisild. The Light Skin Allele of SLC24A5 in South Asians and Europeans Shares Identity by Descent . PLoS Genetics, doi: 10.1371/journal.pgen.1003912. * denotes equal contribution. Full text available.
S.M. Raj, L. Pagani, I. Gallego Romero, T. Kivisild, and W. Amos. A general linear model-based approach for inferring selection to climate. BMC Genetics doi: 10.1186/1471-2156-14-87. Full text available.
S.M. Raj, P. Halebeedu, J.S. Kadandale, M. Mirazón Lahr, I. Gallego Romero, J.R. Yadhav, M. Iliescu, N. Rai, F. Crivellaro, G. Chaubey, R. Villems, K. Thangaraj, K. Muniyappa, H.S. Chandra, and T. Kivisild. Variation at Diabetes- and Obesity-Associated Loci May Mirror Neutral Patterns of Human Population Diversity and Diabetes Prevalence in India. Annals of Human Genetics, doi: 10.1111/ahg.12028. Full text available.
2012
I. Gallego Romero, I Ruvinsky and Y. Gilad. Comparative studies of gene expression and the evolution of gene regulation. Nature Reviews Genetics, doi: 10.1038/nrg3229. Link.
L. Pagani, T. Kivisild, A. Tarekegn, R. Ekong, C. Plaster, I. Gallego Romero, Q. Ayub, S.Q. Mehdi, M.G. Thomas, D. Luiselli, E. Bekele, N. Bradman, D.J. Balding and C. Tyler-Smith. Ethiopian Genetic Diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. American Journal of Human Genetics, doi: 10.1016/j.ajhg.2012.05.015. Full text available.
D.G. MacArthur, S. Balasubramanian, A. Frankish, N. Huang, J. Morris, K. Walter, L. Jostins, L. Habegger, J.K. Pickrell, S.B. Montgomery, C.A. Albers, Z.D. Zhang, D.F. Conrad, G. Lunter, H. Zheng, Q. Ayub, M.A. DePristo, E. Banks, M. Hu, R.E. Handsaker, J.A. Rosenfeld, M. Fromer, M. Jin, X. Jasmine Mu, E. Khurana, K. Ye, M. Kay, G.I. Saunders, M-M. Suner, T. Hunt, I.H.A. Barnes, C. Amid, D.R. Carvalho-Silva, A.H. Bignell, C. Snow, B. Yngvadottir, S. Bumpstead, D.N. Cooper, Y. Xue, I. Gallego Romero, 1000 Genomes Project Consortium, J. Wang, Y. Li, R.A. Gibbs, S.A. McCarroll, E.T. Dermitzakis, J.K. Pritchard, J.C. Barrett, J. Harrow, M.E. Hurles, M.B. Gerstein and C. Tyler-Smith. A Systematic Survey of Loss-of-Function Variants in Human Protein-Coding Genes. Science, doi: 10.1126/science.1215040. Full text available.
2011
M. Hu, Q. Ayub, J.A. Guerra-Assunção, Q. Long, Z. Ning, N. Huang, I. Gallego Romero, L. Mamanova, P. Akan, X. Liu, A.J. Coffey, D.J. Turner, H. Swerdlow, J. Burton, M.A. Quail, D.F. Conrad, A.J. Enright, C. Tyler-Smith and Y. Xue. Exploration of signals of positive selection derived from genotype-based human genome scans using re-sequencing data. Human Genetics, doi: 10.1007/s00439-011-1111-9. Link.
M. Metspalu*, I. Gallego Romero*, B. Yunusbayev*, G. Chaubey, C. Basu Mallick, G. Hudjashov, M. Nelis, R. Mägi, E. Metspalu, M. Remm, R. Pitchappan, L. Singh, K. Thangaraj, R. Villems, and T. Kivisild. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. American Journal of Human Genetics, doi: 10.1016/j.ajhg.2011.11.010. * denotes equal contribution. Full text available.
M. Rasmussen, X. Guo, Y. Wang, K.E. Lohmueller, S. Rasmussen, A. Albrechtsen, L. Skotte, S. Lindgreen, M. Metspalu, T. Jombart, T. Kivisild, W. Zhai, A. Eriksson, A. Manica, L. Orlando, F. De La Vega, S. Tridico, E. Metspalu, K. Nielsen, M.C. Avila-Arcos, J.V. Moreno-Mayar, C. Muller, J. Dortch, M.T. Gilbert, O. Lund, A. Wesolowska, M. Karmin, L.A. Weinert, B. Wang, J. Li, S. Tai, F. Xiao, T. Hanihara, G. van Driem, A.R. Jha, F.X. Ricaut, P. de Knijff, A.B. Migliano, I. Gallego Romero, K. Kristiansen, D.M. Lambert, S. Brunak, P. Forster, B. Brinkmann, O. Nehlich, M. Bunce, M. Richards, R. Gupta, C.D. Bustamante, A. Krogh, R. A. Foley, M.M. Lahr, F. Balloux, T. Sicheritz-Pontén, R. Villems, R. Nielsen, W. Jun and E. Willerslev. An Aboriginal Australian Genome Reveals Separate Human Dispersals into Asia. Science, doi: 10.1126/science.1211177. Full text available.
I. Gallego Romero, C. Basu Mallick, A Liebert, F Crivellaro, G Chaubey, A Liebert, Y Ital, R. Pitchappan, M. Metspalu, M. Eaaswarkanth, R. Villems, L. Singh, K. Thangaraj, M. G. Thomas, D. Swallow, M Mirazón Lahr, and T. Kivisild. Herders of Indian and European cattle share their predominant allele for milk tolerance. Molecular Biology and Evolution, doi: 10.1093/molbev/msr190. Full text available.
2010-2008
G. Chaubey, M. Metspalu, Y. Choi, R. Maagi, I. Gallego Romero, P. Soares, M. van Oven, D. M. Behar, S. Rootsi, G. Hudjashov, C. Basu Mallick, M. Karmin, M. Nelis, J. Parik, A. G. Reddy, E. Metspalu, G. van Driem, Y. Xue, C. Tyler-Smith, K. Thangaraj, L. Singh, M. Remm, M. B. Richards, M. Mirazón Lahr, M. Kayser, R. Villems, and T. Kivisild. Population genetic structure in Indian Austroasiatic speakers: The role of landscape barriers and sex-specific admixture. Molecular Biology and Evolution, doi: 10.1093/molbev/msq288, 2010. Full text available.
M. Eaaswarkhanth, I. Haque, Z. Ravesh, I. Gallego Romero, P. R. Meganathan, B. Dubey, F. A. Khan, G. Chaubey, T. Kivisild, C. Tyler-Smith, L. Singh, and K. Thangaraj. Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations. European Journal of Human Genetics, 18(3):354-63, 2010. Full text available.
I. Gallego Romero, A. Manica, J. Goudet, L. J. Lawson-Handley, and F. Balloux. How accurate is the current picture of human genetic variation? Heredity, 102(2):120-6, 2009. Full text available.
I. Gallego Romero and C. Ober. CFTR mutations and reproductive outcomes in a population isolate. Human Genetics, 122(6):583-8, 2008. Full text available.
The lab is based in the Melbourne Integrative Genomics unit at the University of Melbourne. We are part of the School of BioSciences, but also enjoy really close ties to the Centre for Stem Cell Systems, where Irene is an associate member, and with whom we share our wet laboratory research facilities.
Our physical address is:
Gallego Romero Lab,
Melbourne Integrative Genomics
Building 184
University of Melbourne
Parkville, VIC, 3013
Australia


