Our Research
The lab is focused on human evolution, especially in the ways in which gene regulatory processes have contributed to it. Because gene regulation is not easily predicted (at least for now!) from sequence-level data, we use induced pluripotent stem cells (iPSCs) as models to understand the means by which multiple cellular mechanisms interact to regulate gene expression, and, under the action of natural selection, ultimately give rise to inter-species or population-level differences. We are one of few labs worldwide with an established record in generating iPSC lines from non-model organisms, and we combine this expertise with our experience working with the latest genomic technologies.
At the University of Melbourne we enjoy very close links with the School of BioSciences and of Mathematics and Statistics, as well as the Centre for Stem Cell Systems, and we are affiliate members of Stem Cells Australia, a multi-institution, multi-disciplinary initiative that brings together Australia’s leading stem cell research groups across different expertises and interests. Further afield, we also have links to researchers at institutions like MCRI, A*STAR, Massey University, Nanyang Technological University, Monash Malaysia, the Wellcome Trust Sanger Institute, Penn State University, the Centro de Regulación Genómica in Barcelona and more.
Our lab is committed to carrying out high quality reproducible research, be that computational or molecular biology based. To that end we make source code and all data and protocols available on publication
Below you can find brief descriptions of some of our research interests.
The role of gene regulatory changes in human evolution
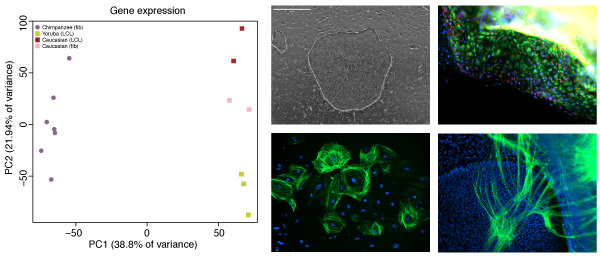
The majority of DNA differences between humans and chimpanzee lie outside protein coding regions of the genome, strongly arguing for a large role of gene regulation as a driver of evolutionary change, especially during embryonic development. However, our understanding of the mechanisms of gene regulation is far from complete, and without access to the right tissue type, it is very difficult to predict the consequences of any mutation. Working with iPSCs gives us access to the right cell type, at the right developmental time point, from the right animal, and enables us to understand the role these differences play in giving rise to particular phenotypes.
Key publications:
Gallego Romero*, Pavlovic* et al, eLife, 2015.
Gallego Romero, Gilad and Ruvinski, Nat Rev Genetics, 2012.
Local adaptations in human populations
The iPSC-driven comparative evolutionary paradigm can also be pertinent to questions about local adaptation in human groups. Of particular interest to us are populations living in the Arctic Circle, who have had to adapt to life in extreme cold conditions, populations adapted to life at high altitude in both Asia and South America, as well as populations living in Island South East Asia, who have been traditionally under-studied. Although there is well-documented evidence for local adaptation in all cases, the functional mechanisms remain elusive. As above, our goal is to combine readily available allelic and sequence information with iPSCs generated from these groups and control populations in order to investigate the actual consequences of these sequence differences in living cells. However, our focus here is not the identification of developmental differences between groups, but rather the identification of population-level functional differences in terminal cell types.
Key publications:
Gallego Romero et al, MBE, 2011.
Metspalu*, Gallego Romero*, Yunushbaev* et al, AJHG, 2011.
Better methods for complex RNA-seq
The use of RNA-seq is widespread. But many experiments – those that depend on field samples, or limited input, or atypical comparisons, suffer from biases that are not often discussed or explored at great depth. We are interested in quantifying and correcting for the effect of these confounders, and in developing analytical approaches that can attenuate these effects. Presently we are most keenly interested in developing robust methods to extensively and affordably perform inter-species RNA-seq comparisons.
Key publications:
Gallego Romero et al, BMC Biology, 2014.